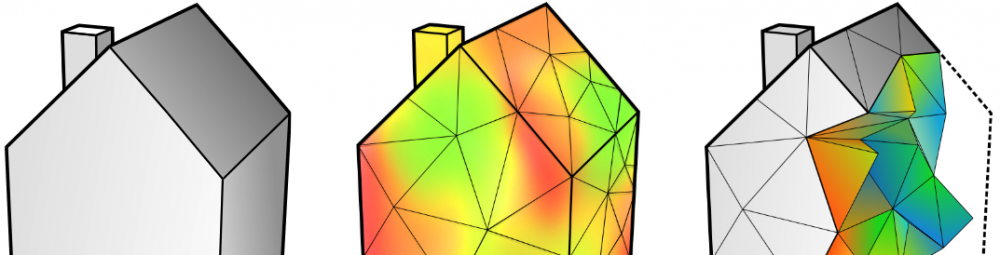
Designing volumetric propertiea is quite a difficult task in the current software culture. None of the standard CAD programs work with volumetric specifications (in my limited experience).
Gladly there is Autodesk Monolith. You can import an STL, which is converted into a voxel field with densities of 100% inside the STL and 0% outside.
You can then model your properties by painting the voxel colors; you need to uncheck geometry so that you only affect the voxel colors, not the densities.
Set the visualization alpha really low so that you can easily see what’s going on inside the model.
When you’re ready go to the rasterization tab and select to only export the material ratio. You can then export the voxel model to a sequence of images. The image lightness encodes the material ratio and the alpha channel encodes the material density.
This is a really nice tool if you want to draw volumetric material property specifications. It can be used to specify an image sequence to be used in Cura as a density voxel model for 3d printing with FDM.