Cura now supports voxel models, by loading in a sequence of images if the first image ends in a number.
The XY are stretched so that the object fits inside while the XY ratio is maintained.
The Z of the images is independently stretched so that all images cover the whole 3D model.
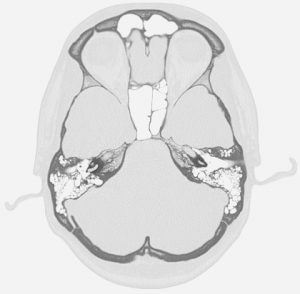
I’ve used it to create some prints which are based on DICOM files from CT-scans.
Note that the surface model is extracted from the DICOM files in a separate program (3D slicer).
I converted the DICOM files into standard png images using MATLAB.
Then I converted the images such that the highest density equaled the density I had set in Cura and a cropped them to where the surface model reached.
This is the result:

This is a sample MATLAB code:
files = dir('osseux/*.dcm');
for file = files'
path = strcat(file.folder, "/", file.name);
a = dicomread(path);
min_val = 300.0 / 32768.0 + .5;
max_val = 1800 / 32768.0 + .5;
minn = min(min(a));
maxx = max(max(a));
a_mapped = uint16(int32(a) + 32768);
adjusted = imadjust(a_mapped, [min_val max_val], [0.0 1.0], 1.2);
[filepath,name,ext] = fileparts(file.name);
outfile = strcat(file.folder, '/output/', name, ".png");
imwrite(65535 - adjusted, outfile{1});
end
Don’t blame me if this crappy hacked together code doesn’t work on your machine! 😉
Here’s a sample converted .cdm file: